dat <- dat |>
mutate(
time_potentially_exposed = runif(n(), 0, 45),
time_potentially_exposed = to_weeks_days(time_potentially_exposed),
time_exposed = ifelse(
time_potentially_exposed < gest_week,
time_potentially_exposed,
NA
),
week_exposed = floor(time_exposed),
ever_exposed = ifelse(!is.na(time_exposed), 1, 0),
# Set time_exposed to arbitrarily high value if never exposed
# to not have to deal with NAs in later analyses
time_exposed = ifelse(is.na(time_exposed), 1000, time_exposed),
week_exposed = ifelse(is.na(week_exposed), 1000, week_exposed)
)Session 1.3
Immortal time bias
Recap
- Not everyone will ultimately be at risk for many outcomes of interest due to competing events
- If we have information on them, there are still causal estimands of interest
- Not everyone will be identified/enroll in a study because they have a competing event before that point, in which case we don’t have information on them (left truncation)
- When this is random with respect to the exposure and outcome, we can correct for it
- When it is differential with respect to the exposure, we need to think more carefully…
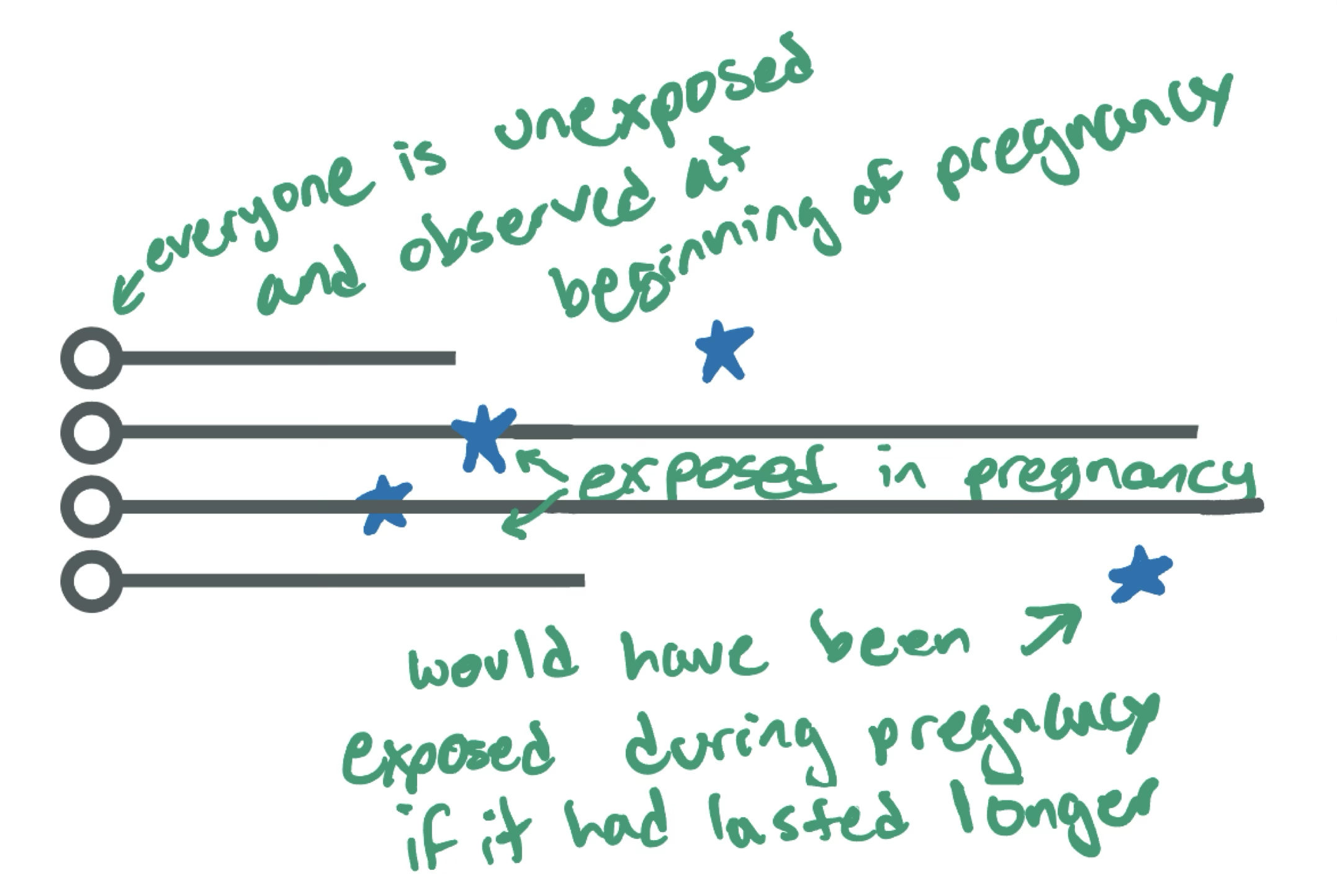
Now let’s think about an exposure that doesn’t occur at the same time for everyone
e.g., vaccination during pregnancy
- It’s still a one-time treatment (let’s assume for simplicity!)
- But it can happen at different times during pregnancy for different people
Again we will generate it under the sharp null hypothesis
Randomly assigned exposure time
The code is explained more in the exercises, but basically:
- Everyone has a random time when they could be exposed (uniformly distributed between 0 and 45 weeks)
- If that time is before the pregnancy ends, they are considered exposed at that time
- If that time is after the pregnancy ends, they are considered never exposed
We are still generating data under the null hypothesis (no effect of exposure on outcome for any individual)
Let’s look at some summary statistics
| Variable | Unexposed N = 3,0481 | Exposed N = 6,9521 | Total N = 10,0001 |
|---|---|---|---|
| Spontaneous abortion (<20 weeks gestation) | |||
| 0 | 1,039 (34%) | 6,365 (92%) | 7,404 (74%) |
| 1 | 2,009 (66%) | 587 (8.4%) | 2,596 (26%) |
| Child born preterm or less than 37 weeks as yes and no | |||
| 0 | 783 (75%) | 5,718 (90%) | 6,501 (88%) |
| 1 | 256 (25%) | 647 (10%) | 903 (12%) |
| Unknown | 2,009 | 587 | 2,596 |
| 1 n (%) | |||
Wait…
I said exposure could happen throughout pregnancy, but spontaneous abortion (SAB) can only happen before 20 weeks
Restricting to a relevant exposure window doesn’t solve the problem
| Variable | Unexposed N = 3,0481 | Exposed before 20 weeks N = 3,8271 | Total N = 6,8751 |
|---|---|---|---|
| Spontaneous abortion (<20 weeks gestation) | |||
| 0 | 1,039 (34%) | 3,240 (85%) | 4,279 (62%) |
| 1 | 2,009 (66%) | 587 (15%) | 2,596 (38%) |
| Child born preterm or less than 37 weeks as yes and no | |||
| 0 | 783 (75%) | 2,847 (88%) | 3,630 (85%) |
| 1 | 256 (25%) | 393 (12%) | 649 (15%) |
| Unknown | 2,009 | 587 | 2,596 |
| 1 n (%) | |||
Shorter pregnancies are less likely to be exposed
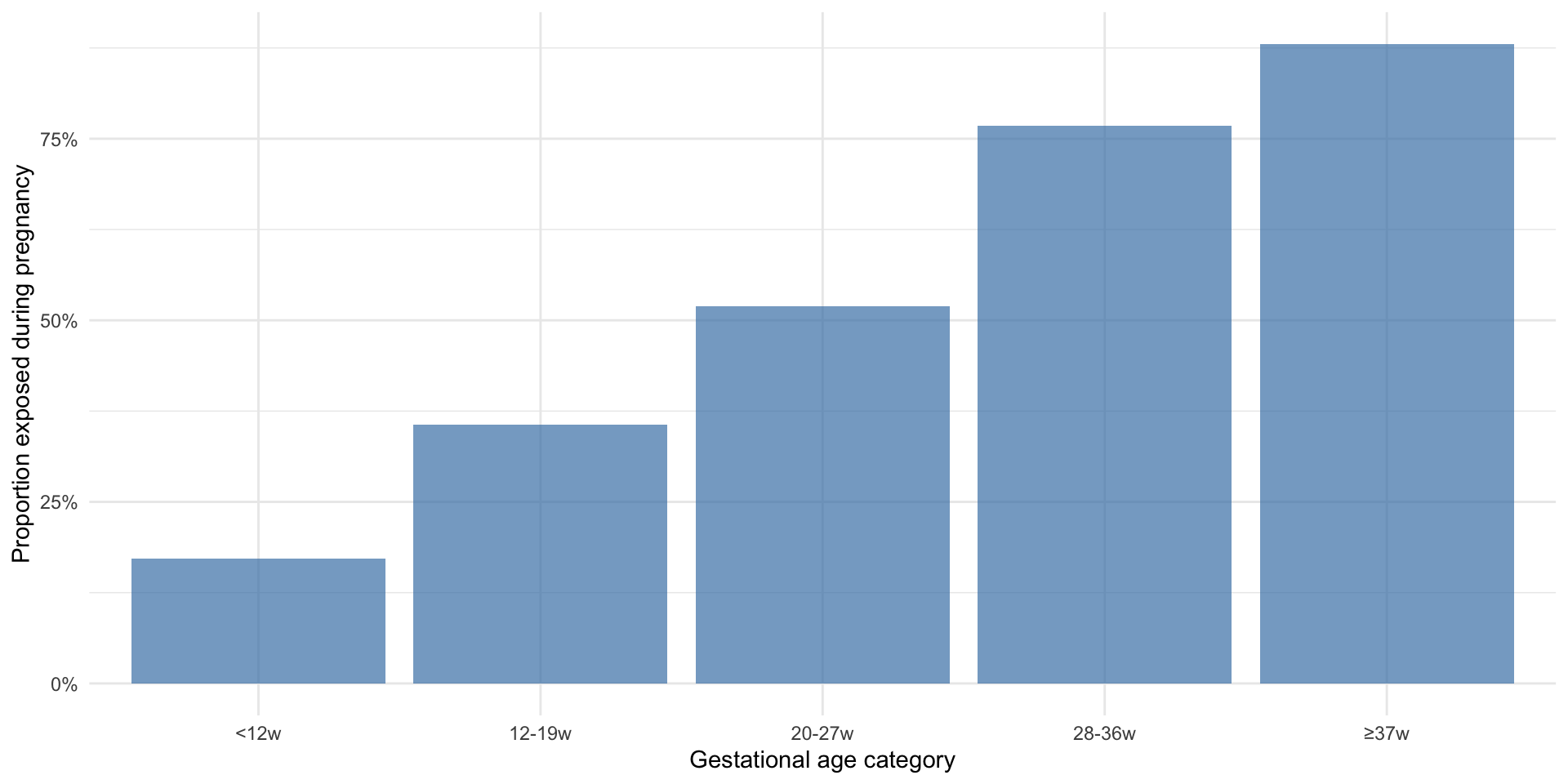
Shorter pregnancies are less likely to be exposed

The bias
- People who are exposed have by definition a pregnancy that lasted long enough to be exposed (whenever that is)
- So exposed group is more likely to be “immortal” (cannot have the outcome) during the time before exposure
- Simpler examples of immortal time bias might compare, e.g., no treatment vs. two years of treatment (must survive that long)
Immortal time bias
Suissa (2008) is probably the most commonly cited description
Several recent papers attempting to describe it structurally, including Mansournia, Nazemipour, and Etminan (2021); Yang, Burgess, and Schooling (2025); Hernán et al. (2025)
Immortal time bias
The term “immortal time bias” suggests that the source of the bias is the immortal time, but it is selection or misclassification that generates the immortal time, leading to bias.
- The difference is whether observations not surviving the “immortal time” are preferentially excluded (selection), or just classified as unexposed (misclassification)
Immortal time biases in pregnancy
- Selection (“post-assignment eligibility”)
- e.g., restricting to people with pregnancies lasting at least 20 weeks, when exposure can happen earlier (i.e., had a competing event before joining the study – left truncation)
- this is a problem whether you start follow-up at 20 weeks or look back and start it earlier
- (this is not a problem if your exposure happens after 20 weeks)
- e.g., restricting to people with pregnancies lasting at least 20 weeks, when exposure can happen earlier (i.e., had a competing event before joining the study – left truncation)
- Misclassification (“post-eligibility assignment”)
- e.g., classifying pregnancies as exposed after surviving to exposure, even though exposure didn’t occur at baseline
“assignment” here refers to treatment assignment, or exposure timing
Immortal time due to selection
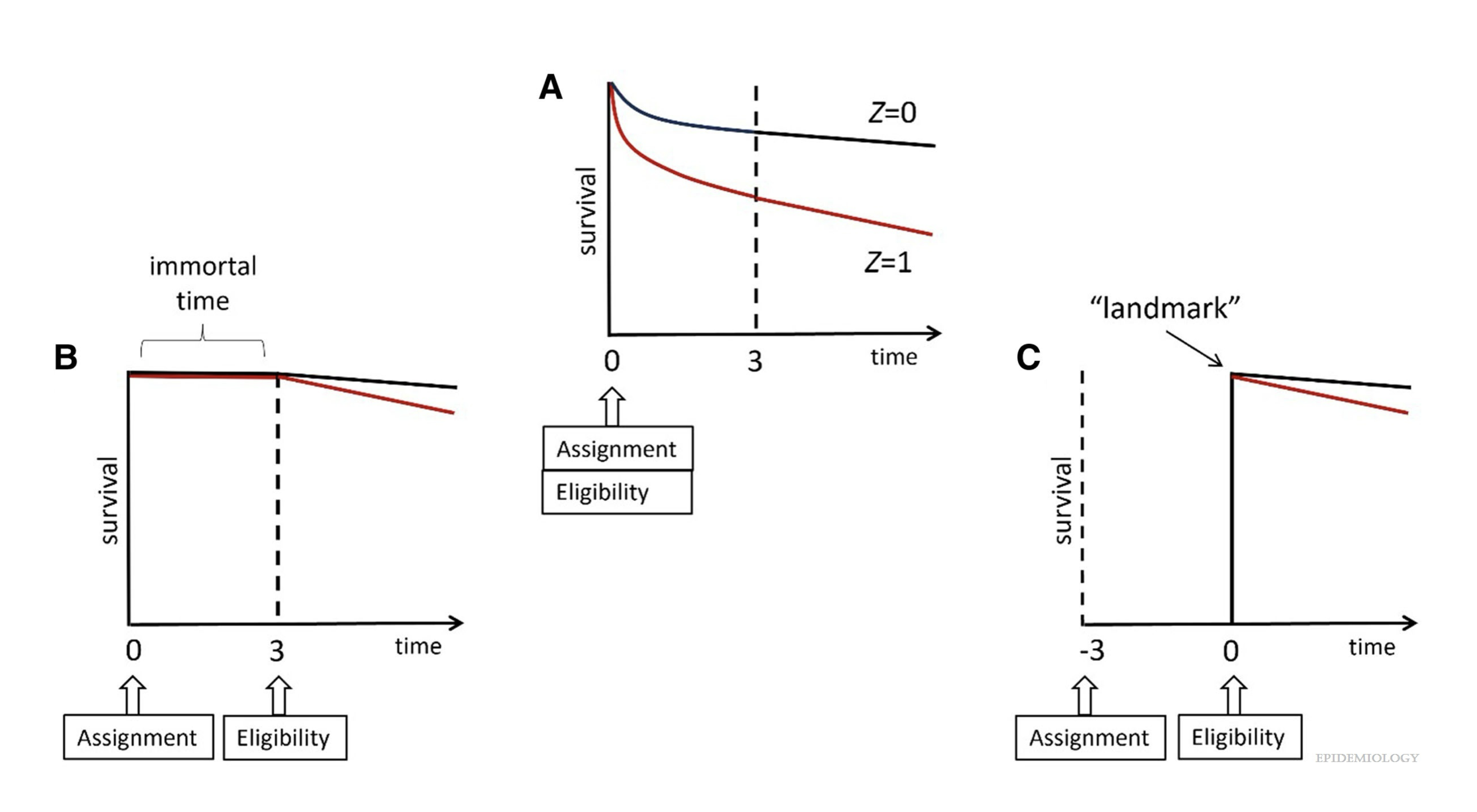
Survival curves from a follow-up study based on all data (A), on data restricted to those who survive at least 3 months with follow-up starting at assignment (B), and on data restricted to those who survive at least 3 months with follow-up starting at 3 months (C). (Hernán et al. 2025)
Immortal time due to misclassification
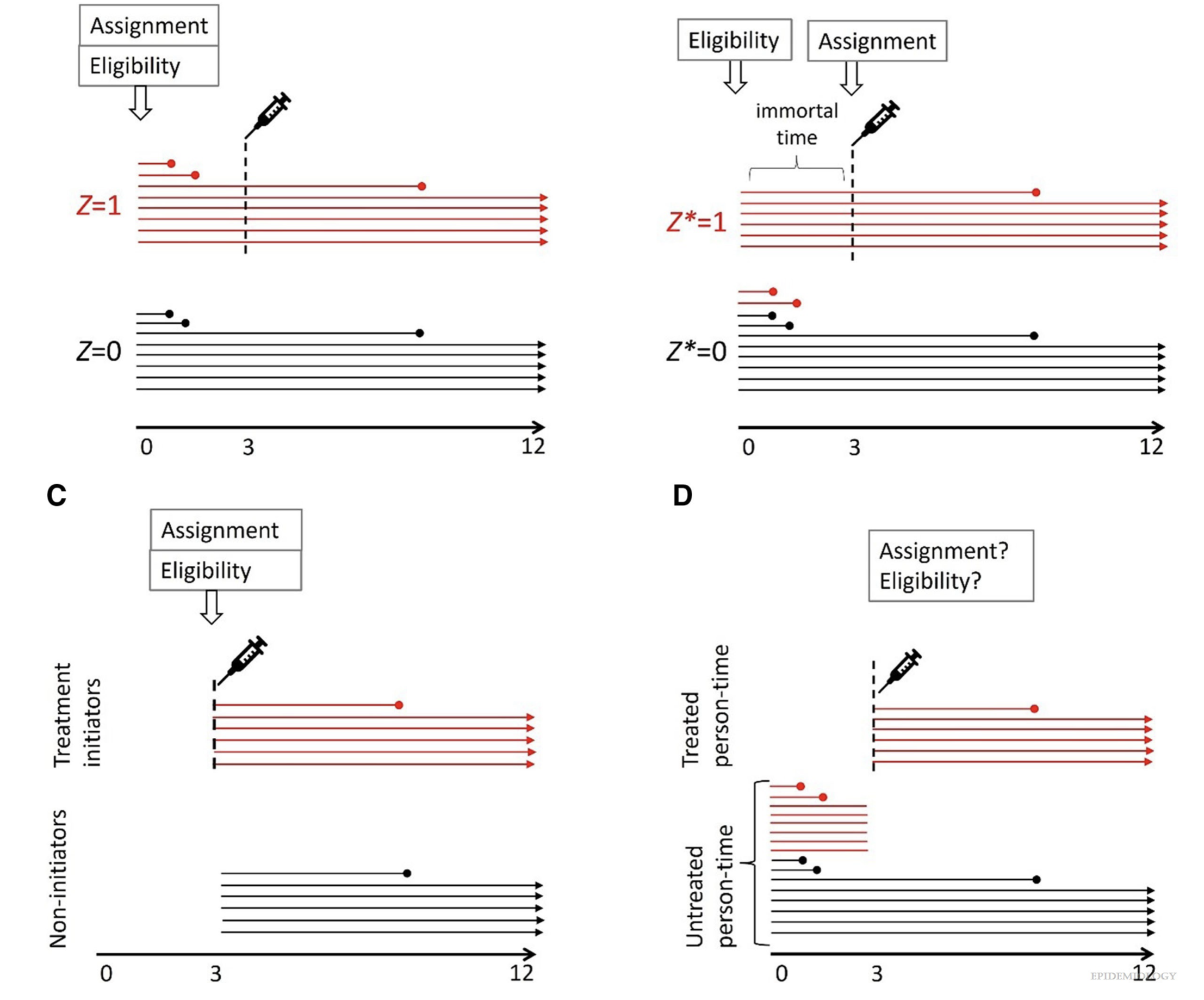
Schematic representation of a study with 16 individuals (horizontal lines) assigned to one of two strategies indistinguishable at time zero: (A) original assignment Z, (B) reconstructed assignment Z* with immortal time, (C) reconfiguration of the data to emulate a study in which individuals are assigned to strategies that are distinguishable at time zero (no immortal time), and (D) reconfiguration of the data to allocate person-time to “exposed” and “unexposed” categories (no immortal time). (Hernán et al. 2025)
What if we are only interested in exposed pregnancies and we have everyone’s data?
We saw that left truncation was a problem when pregnancies had already ended before enrolling or being identified in a study
- The same thing happens even if everyone is enrolled at the start of pregnancy, but we are comparing exposed pregnancies to each other based on timing of exposure
This means that we can’t even tell whether there is an effect of timing of exposure
Let’s look at what the risk of SAB is by week of exposure
The risks are variable, but as expected, they decrease with gestational age
Remember that we generated data under the null hypothesis (no effect of exposure on outcome for any individual)
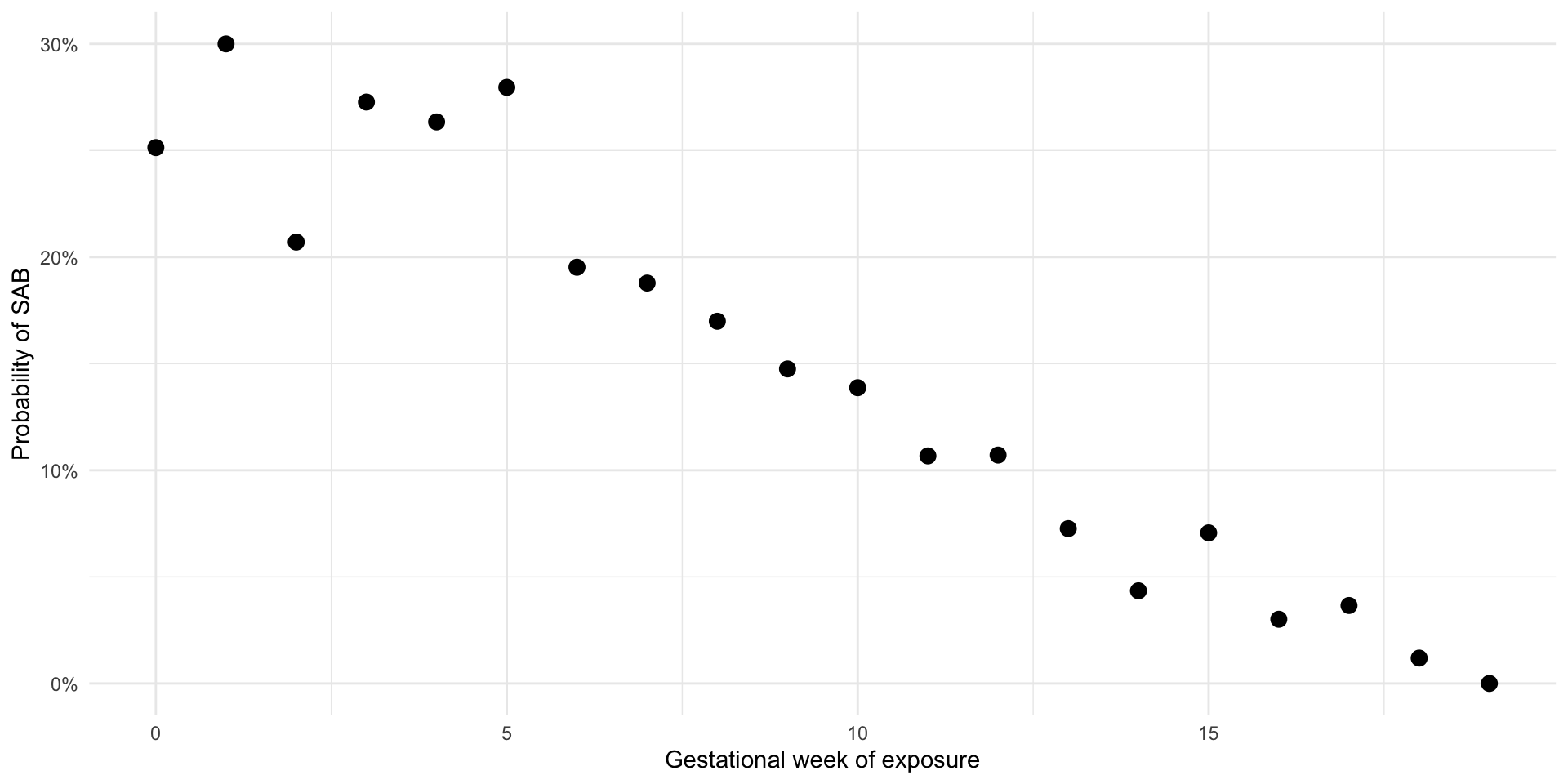
Cumulative incidence of SAB by week of exposure
What if we compare exposed to unexposed but still pregnant?
- We can compare pregnancies exposed in week 6 to those still pregnant but unexposed in week 6
- We can compare pregnancies exposed in week 7 to those still pregnant but unexposed in week 7
- This will include most of the previous comparison group
- And so on…
Let’s think about what this looks like
Who will be in each group for a comparison at week 6? Who will be in each group for a comparison at week 7?
| ID | week_exposed | gest_week |
|---|---|---|
| 1 | 6 | 34 |
| 2 | 7 | 18 |
| 3 | 32 | 40 |
| 4 | 1000 | 6.5 |
| 5 | 1000 | 36 |
Recall that we have set week_exposed to 1000 for unexposed pregnancies
Comparisons
| ID | week_exposed | gest_week |
|---|---|---|
| 1 | 6 | 34 |
| 2 | 7 | 18 |
| 3 | 32 | 40 |
| 4 | 1000 | 6.5 |
| 5 | 1000 | 36 |
- At week 6:
- Exposed: ID 1
- Unexposed but still pregnant: ID 2, 3, 4, 5
- At week 7:
- Exposed: ID 2
- Unexposed but still pregnant: ID 3, 5
How do we implement this in code?
- Create a dataset for each week of pregnancy
- In each dataset, classify people as exposed if they were exposed that week, unexposed if they are still pregnant but unexposed that week, and exclude if they already had the outcome or the pregnancy ended
Let’s do this for all weeks
We’ll use the crossing() function to create a dataset for each week of pregnancy (all concatenated together)
sab_weekly <- dat |>
crossing(week_comparison = 6:19)
sab_weekly |>
select(ID, week_exposed, gest_week, week_comparison)# A tibble: 140,000 × 4
ID week_exposed gest_week week_comparison
<int> <dbl> <dbl> <int>
1 1 7 40.6 6
2 1 7 40.6 7
3 1 7 40.6 8
4 1 7 40.6 9
5 1 7 40.6 10
6 1 7 40.6 11
7 1 7 40.6 12
8 1 7 40.6 13
9 1 7 40.6 14
10 1 7 40.6 15
# ℹ 139,990 more rowsLet’s do this for all weeks
Now we can remove 1) those who already had an event, and 2) those who already had an exposure (so they are either exposed that week, or still unexposed)
sab_weekly <- sab_weekly |>
# still pregnant at trial time
filter(gest_week >= week_comparison) |>
filter(
# eligible at trial time (removing anyone exposed before the trial)
week_exposed > week_comparison | # exposed never or later
week_exposed == week_comparison # exposed now
) |>
mutate(
# indicate whether exposed this week
exposed_now = ifelse(week_exposed == week_comparison, 1, 0)
)Example individual
| ID | week_exposed | gest_week | week_comparison | exposed_now |
|---|---|---|---|---|
| 10281 | 7 | 38 | 6 | 0 |
| 10281 | 7 | 38 | 7 | 1 |
Example individual
| ID | week_exposed | gest_week | week_comparison | exposed_now |
|---|---|---|---|---|
| 26859 | 1000 | 8 | 6 | 0 |
| 26859 | 1000 | 8 | 7 | 0 |
| 26859 | 1000 | 8 | 8 | 0 |
Example individual
| ID | week_exposed | gest_week | week_comparison | exposed_now |
|---|---|---|---|---|
| 2589 | 1000 | 38 | 6 | 0 |
| 2589 | 1000 | 38 | 7 | 0 |
| 2589 | 1000 | 38 | 8 | 0 |
| 2589 | 1000 | 38 | 9 | 0 |
| 2589 | 1000 | 38 | 10 | 0 |
| 2589 | 1000 | 38 | 11 | 0 |
| 2589 | 1000 | 38 | 12 | 0 |
| 2589 | 1000 | 38 | 13 | 0 |
| 2589 | 1000 | 38 | 14 | 0 |
| 2589 | 1000 | 38 | 15 | 0 |
| 2589 | 1000 | 38 | 16 | 0 |
| 2589 | 1000 | 38 | 17 | 0 |
| 2589 | 1000 | 38 | 18 | 0 |
| 2589 | 1000 | 38 | 19 | 0 |
We can see the expected patterns in sample size across comparisons
| week_comparison | n_0 | n_1 |
|---|---|---|
| 6 | 8169 | 210 |
| 7 | 7689 | 197 |
| 8 | 7247 | 206 |
| 9 | 6871 | 183 |
| 10 | 6514 | 173 |
| 11 | 6182 | 178 |
| 12 | 5895 | 168 |
| 13 | 5617 | 179 |
| 14 | 5342 | 207 |
| 15 | 5082 | 184 |
| 16 | 4862 | 166 |
| 17 | 4636 | 164 |
| 18 | 4411 | 168 |
| 19 | 4209 | 158 |
Now we can compare
# Calculate risks by week
sab_risks_weekly <- sab_weekly |>
group_by(week_comparison, exposed_now) |>
summarise(pr_sab = mean(sab))
sab_risks_weekly# A tibble: 28 × 3
# Groups: week_comparison [14]
week_comparison exposed_now pr_sab
<int> <dbl> <dbl>
1 6 0 0.230
2 6 1 0.195
3 7 0 0.203
4 7 1 0.188
5 8 0 0.178
6 8 1 0.170
7 9 0 0.156
8 9 1 0.148
9 10 0 0.132
10 10 1 0.139
# ℹ 18 more rowsAll comparisons

Believe it or not this is still not an entirely fair comparison!
This will motivate target trial emulation and the clone-censor-weighting approach in the next sessions